EN | PT
 |
 |
 |
 |
_________________________________________
 |
General information - TeamCork oak draft genome - Cork oak F1 populationPublications - Quercus Resources |
 |
_________________________________________
A detailed description of the datasets, analysis pipelines and results obtained for the cork oak draft genome assembly can be found in the manuscript published in the journal Scientific Data (https://www.nature.com/articles/sdata201869).
High-throughput sequence data generated in the Illumina platform was used for the assembly of the cork oak draft genome. A mixture of paired-end and mate-pair libraries were produced, to perform the de novo assembly and scaffolding steps, and a total of 10,560,988,448 reads were generated. RNA-seq data was also produced for five cork oak tissues, including xylem, inner bark, phellem, pollen and leaf. The transcriptomic data (1,530,447,601 reads) was subsequently used in the annotation of the draft genome.
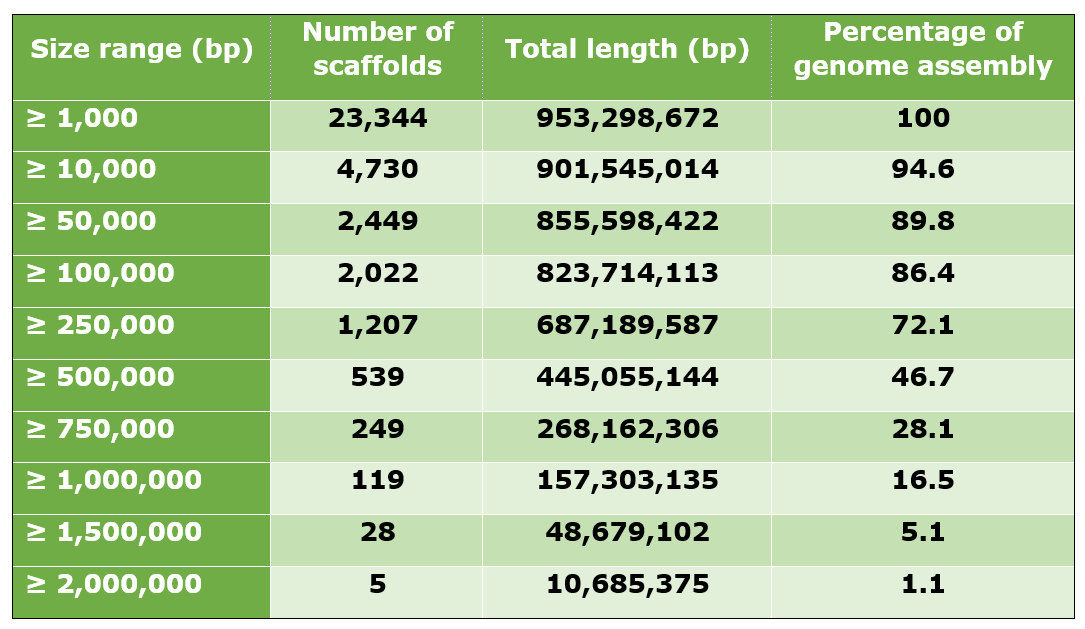
The cork oak draft genome contains 23,344 scaffolds, for an assembly length of 953.3 Mb. The N50 observed was 465.2 Kb, while the longest scaffold was 2,284,287 bp in length. A large percentage of the draft genome was contained in the longer scaffolds. For example, the 2,022 scaffolds with a minimum length of 100 Kb contained 823.7 Mb of the genome, which represented 86.4% of the assembled genome.
Annotation of the draft genome predicted a total of 79,752 genes and 83,814 transcripts, including 33,658 genes validated with RNA-seq data. Functional annotation of the genome was also performed and an InterPro signature assignment was detected for 69,218 transcripts, which represented 82.6% of the total.